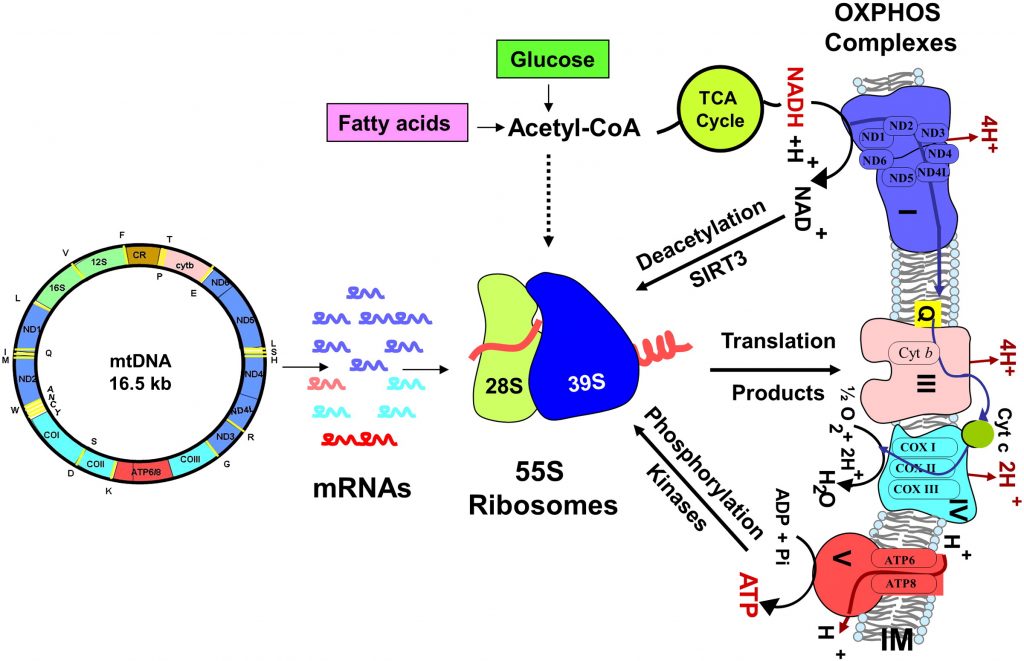
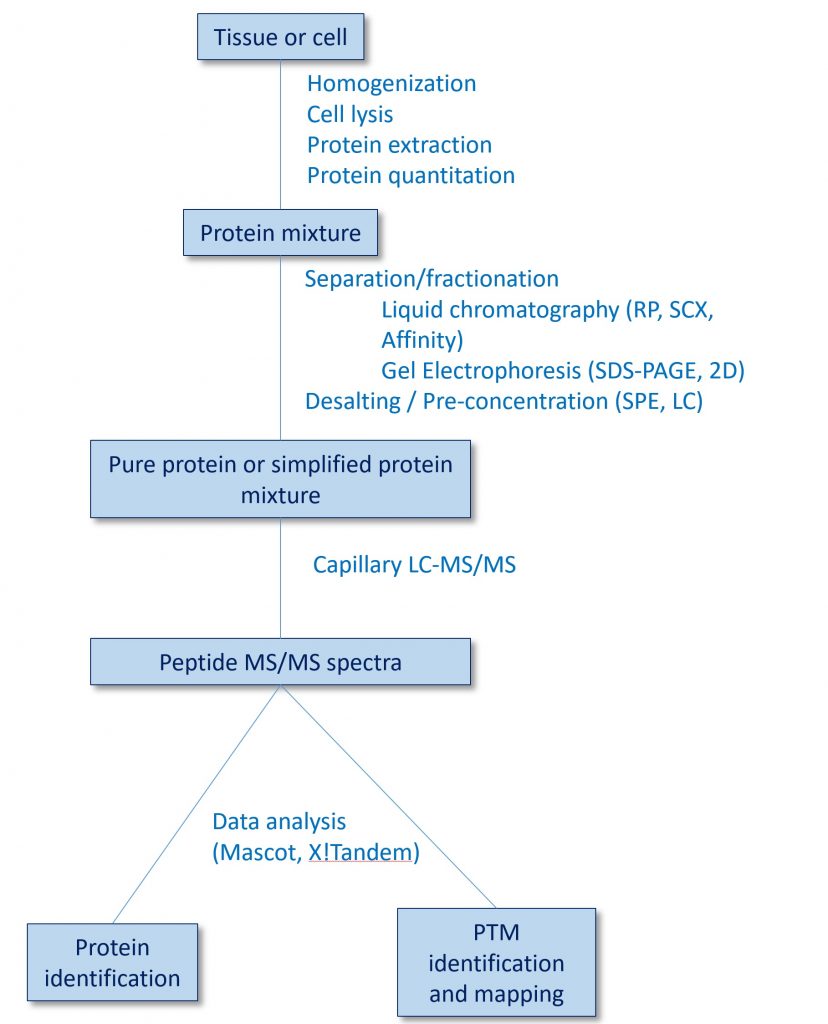
Research in Dr. Koc’s laboratory is focused on understanding how mitochondrial translation is regulated during health and disease using mass spectrometry-based proteomics. As summarized in the above drawing, many players contribute to the energy eventually produced in the inner membrane of the mitochondria. These include circular mitochondrial DNA that encodes 13 proteins of the oxidative phosphorylation process, mitochondrial ribosomes turning these mRNAs into proteins, and the complexes I-V for ATP production. We study the protein components involved in both translation and oxidative phosphorylation using an approach summarized in the below flowchart.
RESOURCES OF KOC LAB
Koc lab has access to the shared resources at the Department of Pharmaceutical Sciences and Research. In addition, we own equipment and resources dedicated to our specific research needs. Here are the major resources that make the research in our lab possible.
- Protein separation techniques
- Gel-based methods such as SDS-PAGE and 2D gel electrophoresis
- Reverse phase and cation exchange (SCX) liquid chromatography
- Affinity techniques to target phosphorylated and acetylated peptides/proteins
- Mass spectrometry sample preparation
- Desalting using reverse-phase cartridges
- Digestion of proteins by proteolytic enzymes such as trypsin either in-gel or in solution
- Fractionation of peptides prior to mass spectrometry
- Capillary liquid chromatography and tandem mass spectrometry
- Protein data analysis and bioinformatics tools

